'a priori' docking
No prior knowledge about the binding sites on your protein is required: only structural data for your ligand and your protein is required.
Run blind docking calculations through a simple interface, without any effort
or prior knwoledge. Check out the advantages of using our platform:

No prior knowledge about the binding sites on your protein is required: only structural data for your ligand and your protein is required.

Save time and power by allowing our platform to carry out the calculations for you.
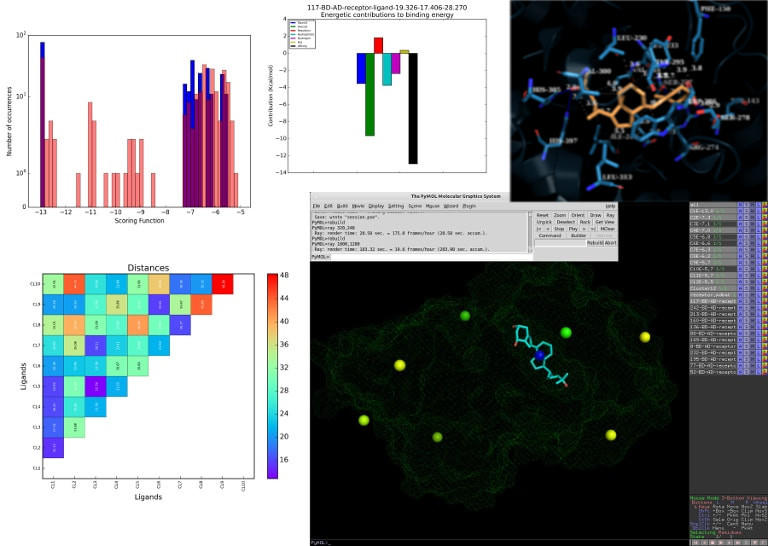
Make your life easier with our advanced features: clustering of overlapping poses, cluster distance and binding energy plots, and a script to easily import the results into PyMOL to better examine the poses and exctrace your own images.
You will be notified whenever one of your calculations is finished, and your results ready for download.
You can preview the results of your calculations online, before downloading any results.
It does not matter if you choose to use our free or premium features. You will always be able to download your data files to handle them yourself.
We are always working on improving the Blind Docking Server, adding new fetures or improving current ones.
Blind Docking server has been used by many academic groups worldwide.
Some of the last citations are:
Ishak, I. H. et al.
Pyrethroid Resistance in Malaysian Populations of Dengue Vector Aedes aegypti Is Mediated by CYP9 Family of Cytochrome P450 Genes. PLoS Negl Trop Dis 11, e0005302–20 (2017).
doi:10.1371/journal.pntd.0005302
Singh, A. & Jana, N. K.
Discovery of potential Zika virus RNA polymerase inhibitors by docking-based virtual screening. Computational Biology and Chemistry 71, 144–151 (2017).
Hassan, N. M., Alhossary, A. A., Mu, Y. & Kwoh, C.-K.
Protein-Ligand Blind Docking Using QuickVina-W With Inter-Process Spatio-Temporal Integration. Sci. Rep. 1–13 (2017).
doi:10.1038/s41598-017-15571-7
Stefania, S. M. et al.
N-thioalkylcarbazoles derivatives as new anti- proliferative agents: synthesis, characterisation and molecular mechanism evaluation. Journal of Enzyme Inhibition and Medicinal Chemistry 0, 434–444 (2018).
Some of our academic users:

Universidad Autónoma de Madrid

CEBAS - CSIC
Our Blind Docking server has been already used by several companies in the pharma and biotech sectors.
Some of our industry users:

Villapharma Research

AQP Ingredients

MD.USE
Supported by